Research
 Adapted from Goldford et al. 2018 Science
Adapted from Goldford et al. 2018 Science
I use laboratory experiments and theory to understand how microbial communities assemble in a given environment, and why.
For this, I use microbial communities from different environments (including the human gut or soil), and combine two approaches: top-down community assembly (to assess what combination of species are selected in a given environment) and bottom-up reconstitution of the dominant species (to ask how they interact in duos, trios,..., in that same environment) (Estrela et al. 2021).
Some of the questions I have asked are:
For this, I use microbial communities from different environments (including the human gut or soil), and combine two approaches: top-down community assembly (to assess what combination of species are selected in a given environment) and bottom-up reconstitution of the dominant species (to ask how they interact in duos, trios,..., in that same environment) (Estrela et al. 2021).
Some of the questions I have asked are:
- Can we predict the influence of nutrient combinations on the composition of microbial communities?
- What are the reproducible and predictable features of microbial communities, which are not, and why?
- Why explains taxonomic variability in microbial community assembly?
- How does migration affect community assembly and dynamics?
- What is the role of species interactions in shaping the emergent properties of these multi-species communities (e.g. function, spatial relationships, and stability)?
 From Estrela et al. 2021 (Cell Systems).
From Estrela et al. 2021 (Cell Systems).
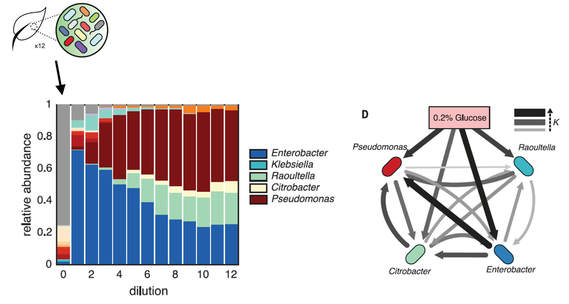
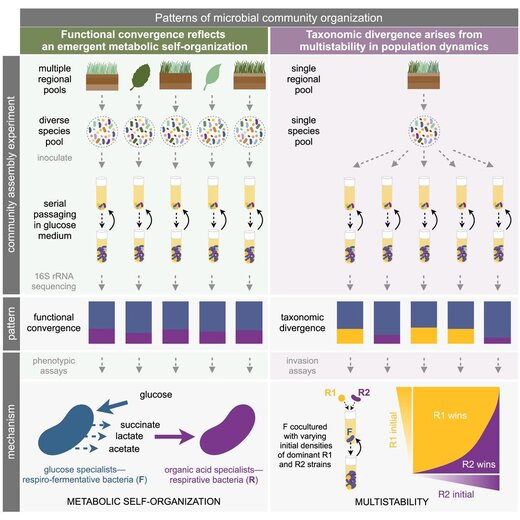
Convergence and divergence in microbial communities
Which descriptive features of microbial communities are reproducible and predictable, which are not, and why? Here we address this question by experimentally studying parallelism and convergence in microbial community assembly in replicate glucose-limited habitats. We show that the previously observed family-level convergence in these habitats reflects a reproducible metabolic organization, where the ratio of the dominant metabolic groups can be explained from a simple resource-partitioning model. In turn, taxonomic divergence among replicate communities arises from multistability in population dynamics. Multistability can also lead to alternative functional states in closed ecosystems but not in metacommunities (Estrela et al. 2021).
|
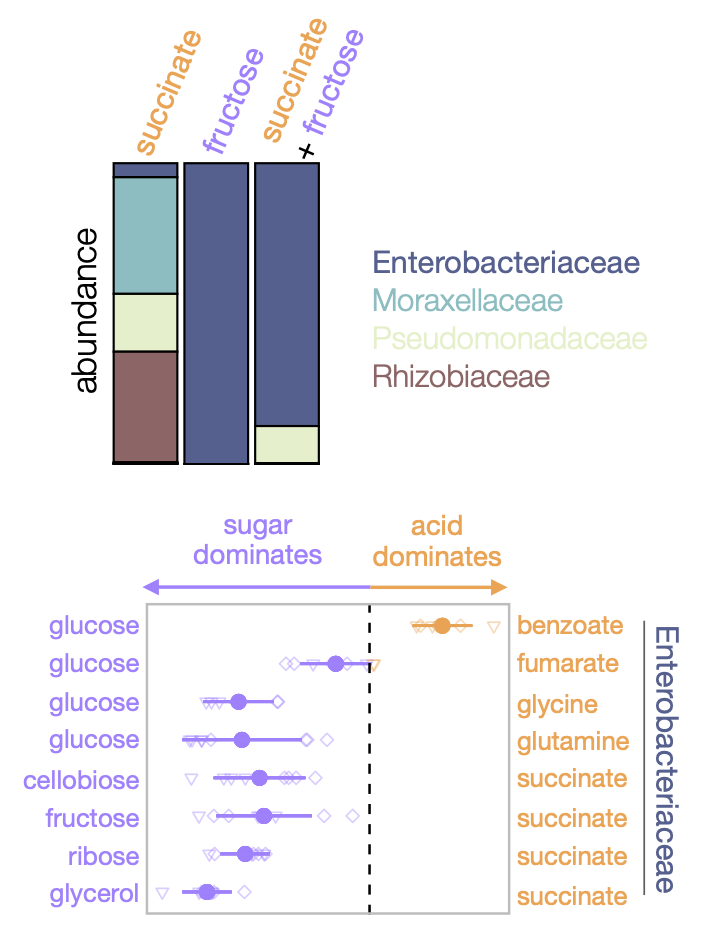
Assembly of microbial communities in mixed nutrient environments
Can we predict how nutrients combine to shape the composition of microbial communities? For instance, if we know what communities assemble in environments with nutrient A and nutrient B separately, can we predict the community that will form in an environment with both nutrients (A+B)? By assembling communities in either pairs of nutrients (sugar-sugar, sugar- organic acid, and organic acid- organic acid) or each nutrient alone, we showed that certain nutrient pairs “interact” in a predictable manner at the family-level of taxonomic organization (Estrela et al. 2021). Specifically, sugars dominate over organic acids. To put it simply, communities assembled in a sugar-acid mixture look more like sugar communities than acid communities. More generally, this reveals that not all nutrients are equal when mixed together, and there exist regularities in how they interact when combined to shape the composition of microbial communities. |
 Adapted from Estrela et al. 2012 (i) and from Estrela & Brown 2013; Estrela et al. 2015 (ii).
Adapted from Estrela et al. 2012 (i) and from Estrela & Brown 2013; Estrela et al. 2015 (ii).
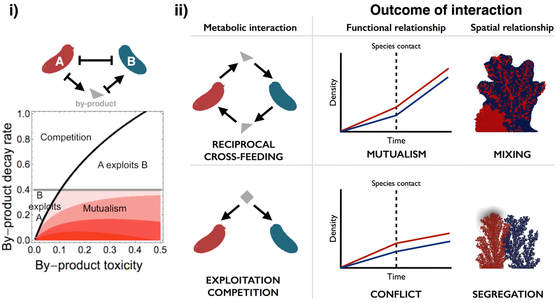
Metabolic interactions and consequences for microbial community functional and spatial relationships
How do metabolic interactions between species (cross-feeding, detoxification) shape their emergent functional and spatial relationships? We have shown that a single mechanism of metabolic exchange - food for detoxification of metabolic waste products- can generate a diverse array of ecological relationships, depending on the properties of the metabolic by-products exchanged (Estrela et al. 2012). In addition, using an individual-based model of biofilm growth, we showed that under competitive interactions microbial species tend to segregate while under mutualistic interactions they tend to mix (Estrela & Brown 2013).
I am also interested in applying fundamental principles of microbial interactions to real world problems, especially health and disease. Applying our findings to the challenge of antibiotic resistance, we recently showed that the ecological and spatial relationships between bacterial species impact how communities respond to antibiotics. In particular, we showed that competition between antibiotic-resistant and sensitive bacteria leads to the 'competitive release' of the resistant type whereas mutualism leads to a ‘mutualistic suppression' where both species are harmed by antibiotics. In addition, these effects may be magnified or dampened, respectively, in structured environments because competitors tend to segregate while mutualists tend to mix as they grow (Estrela & Brown 2018).
How do metabolic interactions between species (cross-feeding, detoxification) shape their emergent functional and spatial relationships? We have shown that a single mechanism of metabolic exchange - food for detoxification of metabolic waste products- can generate a diverse array of ecological relationships, depending on the properties of the metabolic by-products exchanged (Estrela et al. 2012). In addition, using an individual-based model of biofilm growth, we showed that under competitive interactions microbial species tend to segregate while under mutualistic interactions they tend to mix (Estrela & Brown 2013).
I am also interested in applying fundamental principles of microbial interactions to real world problems, especially health and disease. Applying our findings to the challenge of antibiotic resistance, we recently showed that the ecological and spatial relationships between bacterial species impact how communities respond to antibiotics. In particular, we showed that competition between antibiotic-resistant and sensitive bacteria leads to the 'competitive release' of the resistant type whereas mutualism leads to a ‘mutualistic suppression' where both species are harmed by antibiotics. In addition, these effects may be magnified or dampened, respectively, in structured environments because competitors tend to segregate while mutualists tend to mix as they grow (Estrela & Brown 2018).
|
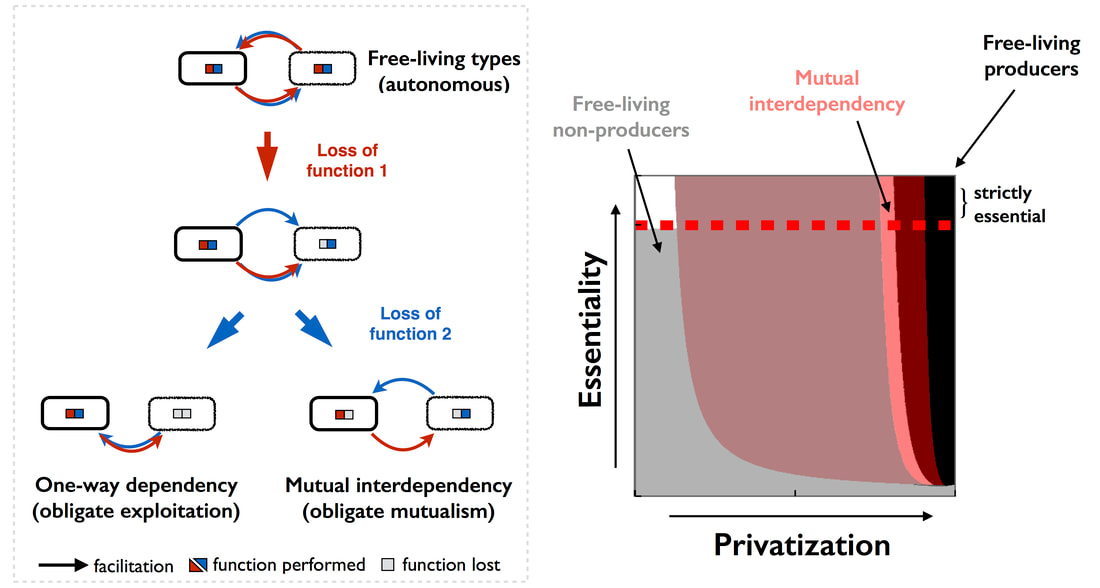
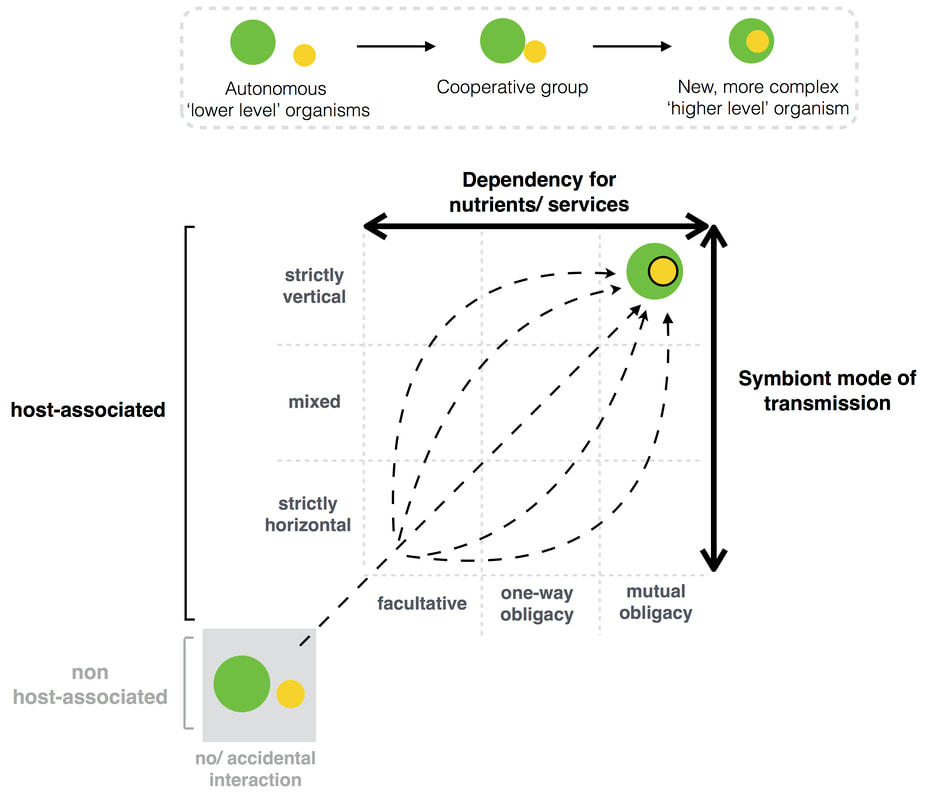
Evolution of microbial dependencies and host-microbe dependencies
Why and how do some species lose functions that leave them dependent on other species? We showed that whether full autonomy, one-way dependency or mutual dependency arises depends on the interplay between costs, essentiality and privatization level of the function (Estrela et al. 2015). In some cases, such dependencies can be so strong that 'individuals' (or organisms) eventually lose their own 'individuality' and merge together, forming a symbiotic organism. I am interested in why and how this happens. That is, why do different species come together to form a single, fully integrated organism (defined as a major egalitarian transition)? What are the evolutionary paths to symbiotic organismality? We have proposed that classifying host-symbiont associations into two interconnected continuum axes: a symbiont’s dependency for dispersal into new hosts (symbiont mode of transmission) and interdependency for reproduction, growth and survival (dependency for nutrients/services) can help us better understand and predict the evolutionary path to symbiotic organismality (Estrela et al. 2016). |